Daniel has been awarded an NSF GRFP to support his research on how biotic and abiotic conditions combine to promote parasite transmission. Well done, Daniel!
Category Archives: Uncategorized
Welcome Daniel & Grant
This semester we welcome Daniel Suh and Grant Foster to the lab. Daniel is starting his PhD in Ecology/IDEAS with interests in parasites and invasive species. Previously, Daniel earned his BS in Biology from Pepperdine and worked as a research assistant in Mike Levy’s zoonotic disease lab at U Penn. Grant is an Ecology/Biology major and honors student. His primary research in the lab is centered on understanding specificity of complex life cycle parasites
Annakate awarded stipend to attend wildlife conservation meeting
Annakate will attend the 73rd Annual Conference of the Southeast Association of Fish and Wildlife Agencies this Fall, after winning a competitive stipend. The meeting will allow Annakate to hear research from state and federal agencies, citizen’s organizations, universities, and private wildlife research groups, fisheries and wildlife scientists, agency enforcement personnel, and other natural resource related organizations. Congratulations Annakate!
Parasite specialism
Andrew led a recent study aimed at characterizing the phylogenetic specialism of around 1500 mammal parasites, finding that patterns of specialism vary among parasite types and transmission modes. Some parasites show an aggregation in the host phylogeny consistent with a leaps-and-creeps history; occasionally, a parasite will take a large leap to a new host species and creep through the phylogeny acquiring related hosts to the one it jumped to. The study provides a vocabulary and context to consider case studies, including zoonoses, and the authors hope that it can catalyze more studies on the evolutionary ecology of parasite host range. The paper is published in Proceedings of the Royal Society B and was profiled on the journals blog.
Estimating the number of mammal species
John was involved in a research project, recently published in Ecology and Evolution, that developed and tested a model to estimate the number of mammal species using taxonomic records.
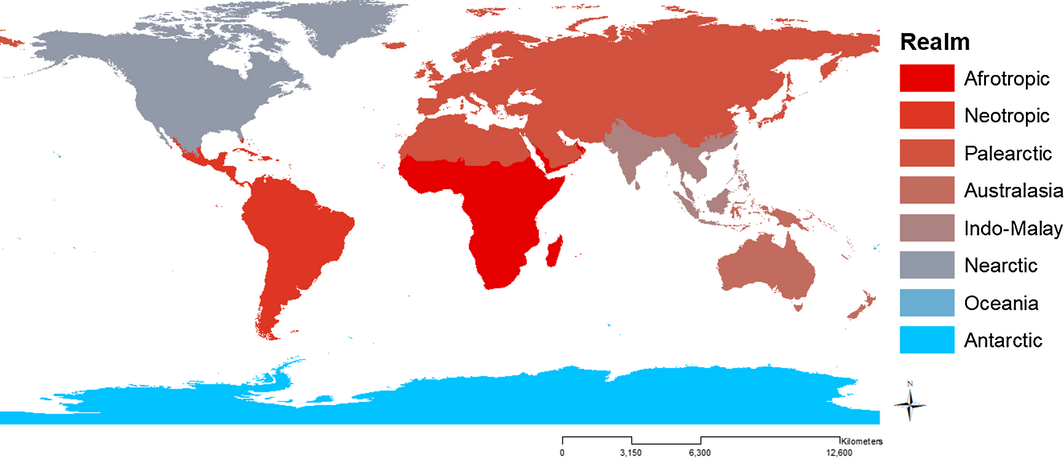
In a study led by Molly Fisher, in collaboration with John Drake and John Gittleman, the group worked to modify and apply a model that used species description records to estimate how many and where mammal species remain to be described. Along with development and application of the model, John worked to develop a simulation, which generated scenarios similar to the real species description records, to test the new model against a previous model. They found that under more complex and realistic scenarios their model outperformed the original model in estimating the total species number. When applying the new model to the real taxonomic data, roughly 5% (303 species) of the mammal species remain to be described. By applying the model to species discovery records for individual biogeographic realms, the group found that the Afrotropics and Neotropics contain the greatest number of undescribed species.
Link prediction
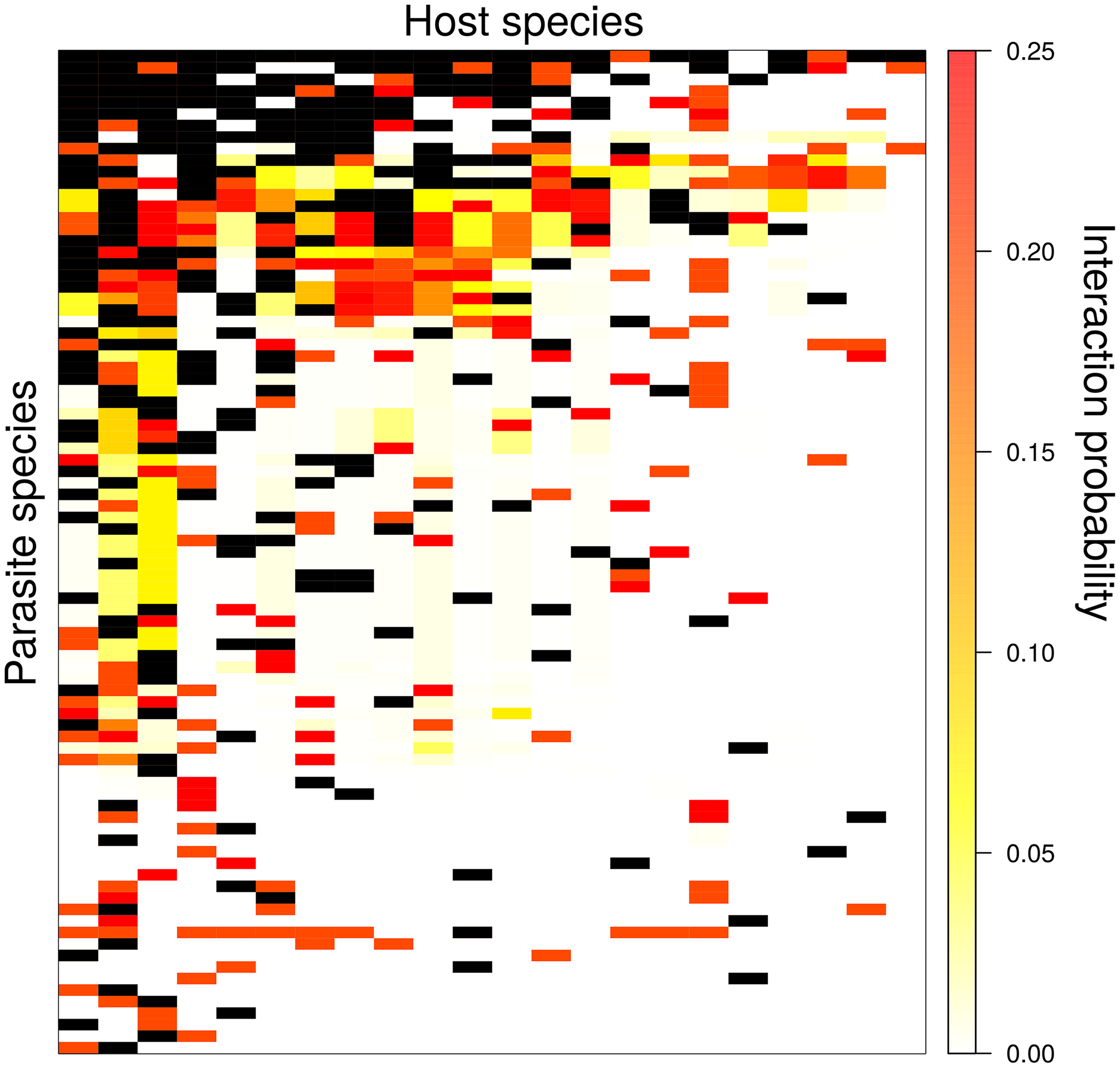
Andrew was involved in a link prediction problem study with recent Odum graduate, Tad Dallas and colleague John Drake, which was recently published in PLoS Computational Biology. They wanted to (i) estimate missing but likely links in a host-parasite network, (ii) test if those missing links changed the network structure and (iii) for a real data set, establish what biological features best predict missing links. They were able to recover randomly removed missing links with high accuracy, and showed that putting these links in to the host-parasite network can dramatically change network structure. For a rodent-parasite dataset, they found that host litter size and diet breadth, along with parasite taxonomy were important features for accurately predicting missing links.
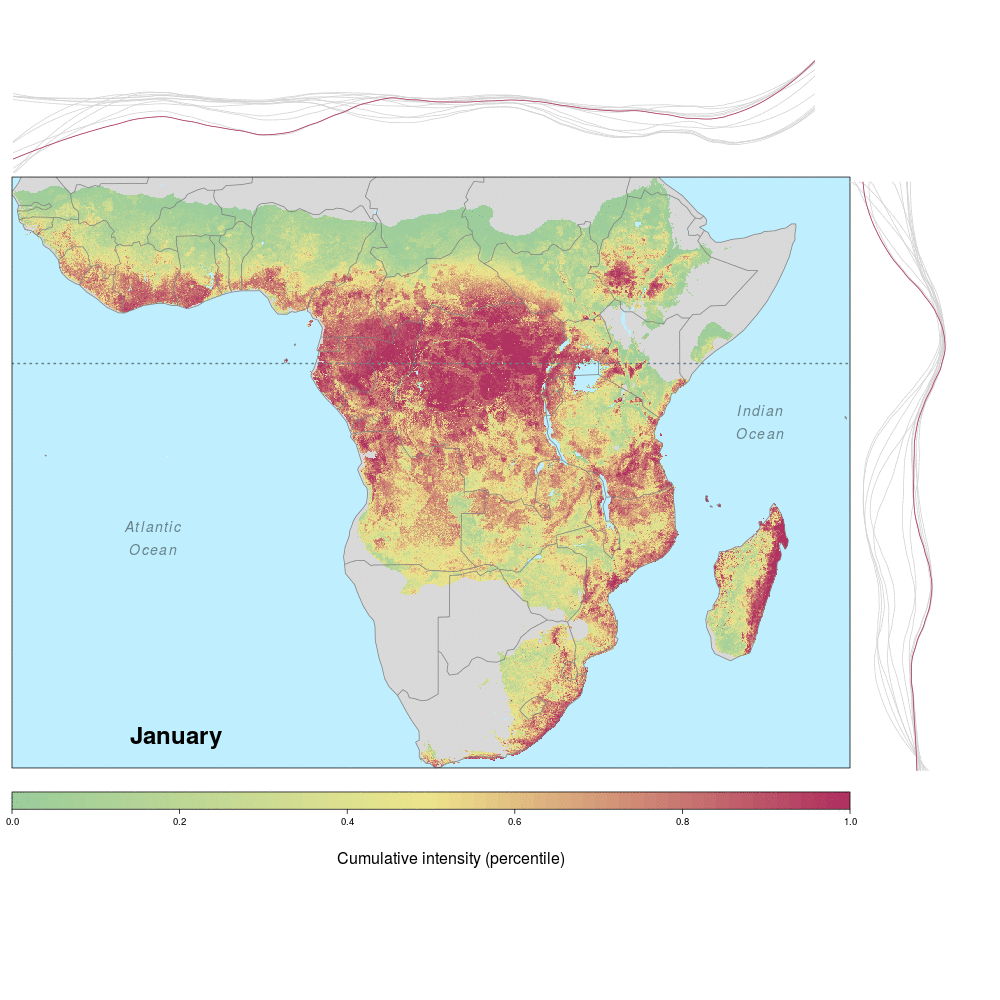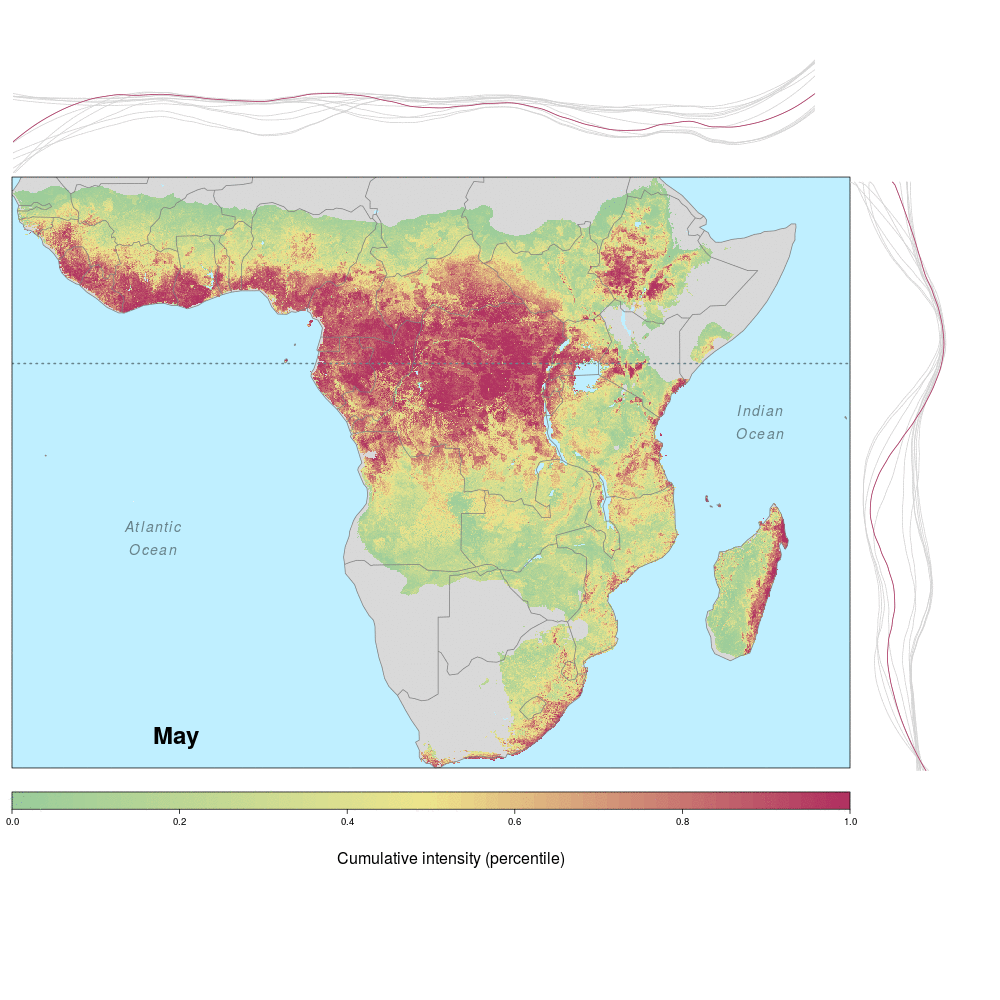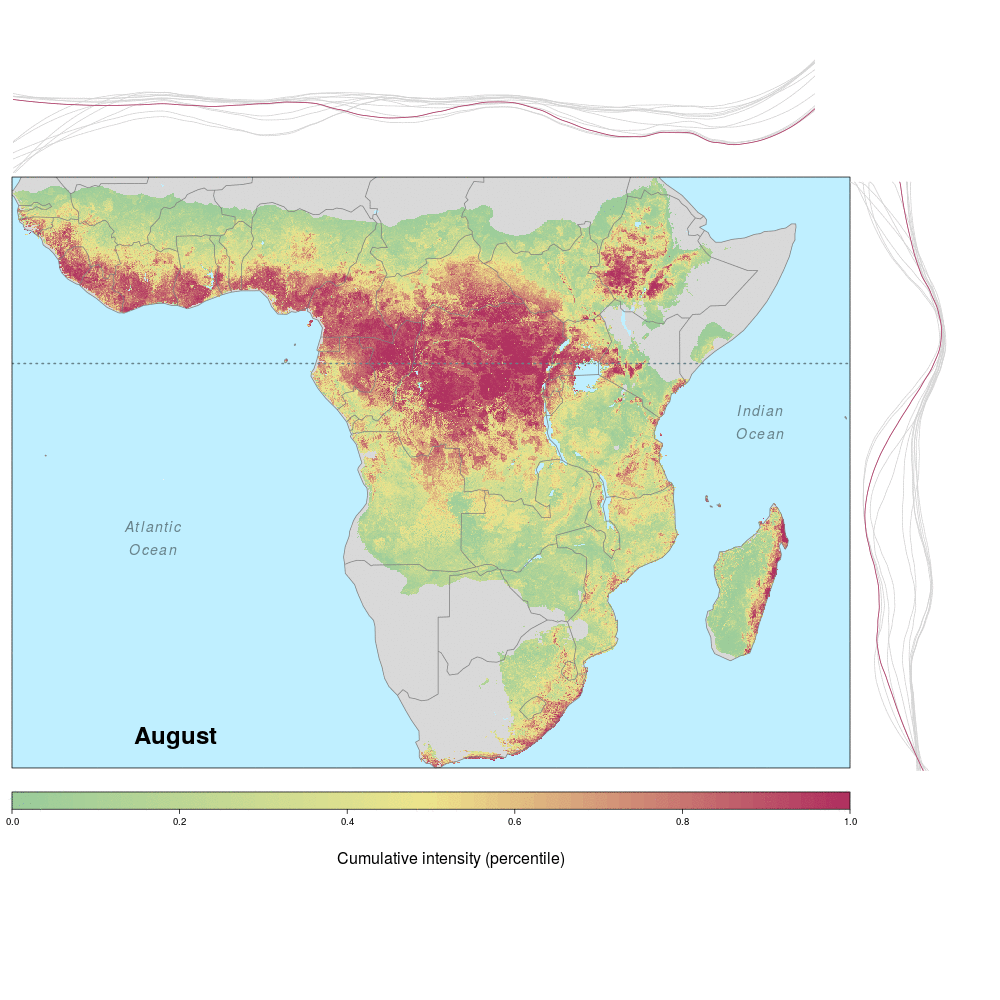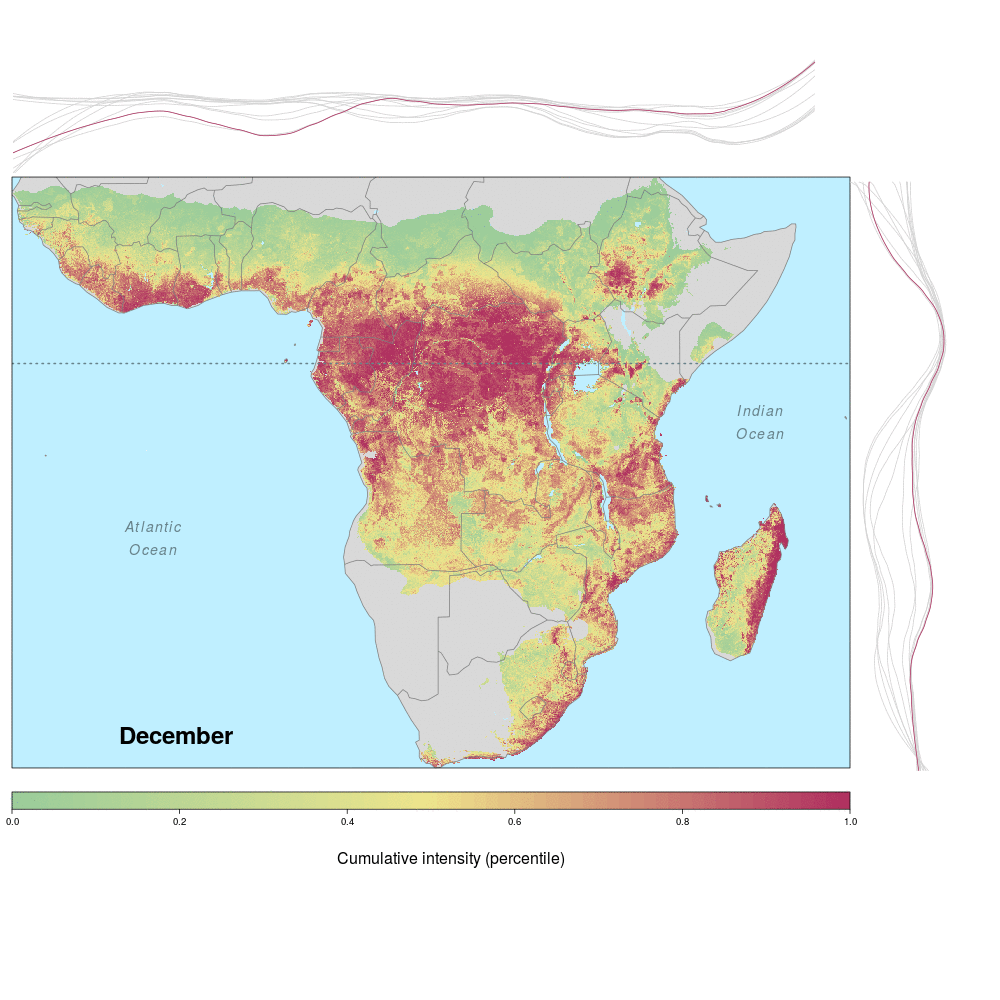
Ebola spillover risk
Andrew and lab alum Laura were involved in a recent study demonstrating the spatio-temporal risk of Ebola spillover in Africa. Using historical spillover data, the research (published in Emerging Infectious Diseases with lead author JP Schmidt of the Odum School of Ecology) shows that much of Central Africa (an historic hotspot) exhibits year round risk, while Southern, Eastern and Western regions of Africa show seasonal risk pulses often associated with transitions between wet and dry seasons. Until the recent 2014 outbreak in West Africa (the worst in history), that area was not considered to be a likely location for Ebola spillover. However, much of Africa, including West Africa, has the right blend of animal and human populations, as well as the climate conditions associated with former outbreaks, at certain times of year.
Jenna awarded summer CURO fellowship and presented at recent CURO symposium
Congratulations to Jenna on being awarded a summer 2017 CURO Fellowship to continue her studies in the lab. Jenna is researching patterns of tick diversity in the mammalian phylogeny to lay the foundations for better understanding the distribution of tick-borne diseases. She recently presented her research, “Patterns of tick-host associations in terrestrial mammals”, at the CURO symposium.
David awarded NSF Graduate Research Fellowship
David Vasquez, co-advised by Vanessa Ezenwa and Andrew Park, is pursuing his doctorate as part of the Interdisciplinary Disease Ecology Across Scales program. His research interests include disease ecology, social network theory, animal behavior, co-infection dynamics, ecoimmunology, and ecophysiology. Congratulations!
Predicting host species
Unsampled host species may associate with parasites of interest, but it can be challenging to predict with which species a given parasite will associate. We tested the predictability of a parasite’s host range (the set of host species it can infect) using a large database of helminth parasites of fish and boosted regression trees (recommended R resource). Tad Dallas led the research, which was recently published in Parasitology. While host traits and environmental variables were predictive, the single best class of predictors was the parasite community itself.